Arguments
Usage instructions
Please follow this link for details on how to install and run this solution.StarDist Model Training Solution for Album
Introduction
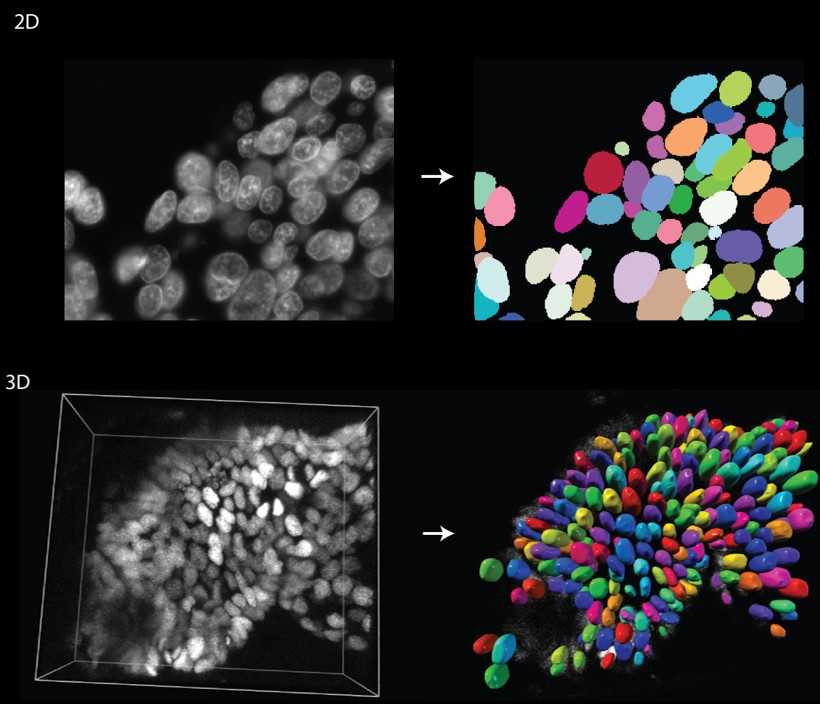
This is an album solution designed to train a StarDist model directly from the command line. The primary goal of this solution is to facilitate the training of models to segment structures using the StarDist approach.
Please refer to the detailed documentation of StarDist for in-depth understanding and guidelines.
Example: Training a 3D StarDist Model for Secretory Granules Segmentation
The purpose of this example is to guide users on training a StarDist model to segment secretory granules from 3D FIB-SEM data. The training procedure and its significance are elaborated in the paper:
Müller, Andreas, et al. "3D FIB-SEM reconstruction of microtubule–organelle interaction in whole primary mouse β cells." Journal of Cell Biology 220.2 (2021).
Dataset Preparation
Download the sample data (or you can prepare your own data in a similar format).
wget https://syncandshare.desy.de/index.php/s/5SJFRtAckjBg5gx/download/data_granules.zip
unzip data_granules.zip
After extracting, your data should be structured as follows:
data_granules
├── train
│ ├── images
│ └── masks
└── val
├── images
└── masks
Installation
Make sure album is already installed. If not, download and install it as described here. Also, don't forget to add the catalog to your album installation, so you can install the solutions from the catalog.
Install the stardist_train and stardist_predict solution by using the graphical user interface (GUI) of album or by running the following command in the terminal:
album install io.github.betaseg:stardist_train:0.1.0
album install io.github.betaseg:stardist_predict:0.1.0
How to use
Training
To start the training process, provide the root directory of the data (using the "root" argument) and the desired output directory (using the "out" argument). By default, the solution will train a 3D stardist model for 100 epochs.
The parameters can be set and run using either the GUI, or by adapting this example for command line usage:
album run stardist_train --root /data/stardist_train/data_granules --out /data/stardist_train/data_granules_out --epochs 10 --steps_per_epoch 15
During training, a TensorBoard will be opened in your browser. You can monitor the training process and the model performance using the TensorBoard. The solution terminates when the training is finished and TensorBoard is closed.
The trained model will be saved in the specified output directory and contains the date and time of the run. The model can be used for inference using the stardist_predict solution.
Predict
For inference, provide the path to an input file (TIF) or directory containing multiple files (TIF) using the `fname_input` argument.
State the `model_name` and its corresponding directory `model_dir`.
The output directory must be specified using the `output_dir` argument.
album run stardist_predict --fname_input /data/stardist_train/data_granules/val/images/high_c1_raw_region_2.tif --model_name 2023_06_28-17_45_35_stardist --model_basedir /data/stardist_train/data_granules_out/ --output_dir /data/stardist_train/predictions
Further documentation:
For further options, parameters and default values, please refer to the info page of the solution:
album info stardist_train
Hardware Requirements
Ensure that your hardware meets the specific requirements of StarDist for efficient training. For training a 3D model, we recommend to use a GPU with at least 8GB of memory.
Citation & License
Stardist is licensed under the BSD 3-Clause License.
If you use this solution, please cite the following paper:
title: Cell Detection with Star-Convex Polygons,
doi: 10.1007/978-3-030-00934-2_30
and
title: Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy,
doi: 10.1109/WACV45572.2020.9093435

