Arguments
Usage instructions
Please follow this link for details on how to install and run this solution.CSBDeep Segmentation Solution for Album
Introduction
This album solution uses the CSBDeep toolbox to train a U-NET model to perfom segmentations from the command line.
The extensive documentation of CSBDeep can be found at http://csbdeep.bioimagecomputing.com/doc/.
This solution consists of two parts:
- CSBDeep-train: This solution is used to train the model
- CSBDeep-predict: This solution is used to perform inference/prediction.
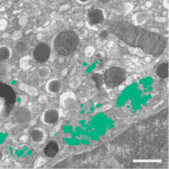
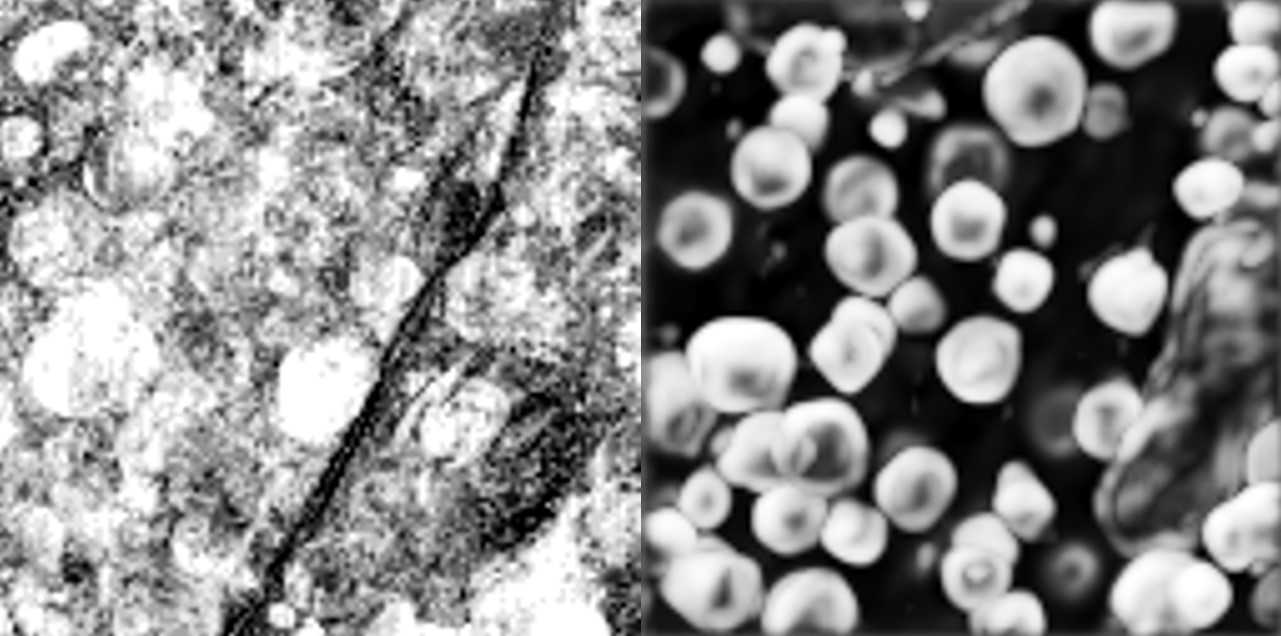
Example: 3D segmentation of golgi aparatus with 3D U-Net
This demonstrates how to use the solution to train a 3D U-Net model to perform semantic segmentation of the golgi aparatus from 3D FIB-SEM data The procedure is described in the paper:
Müller, Andreas, et al. "3D FIB-SEM reconstruction of microtubule–organelle interaction in whole primary mouse β cells." Journal of Cell Biology 220.2 (2021).
Download the example data (or adapt your own data into the same format)
wget https://syncandshare.desy.de/index.php/s/FikPy4k2FHS5L4F/download/data_golgi.zip
unzip data_golgi.zip
which should result in the following folder structure:
data_golgi
├── train
│ ├── images
│ └── masks
└── val
├── images
└── masks
Installation
Make sure album is already installed. If not, download and install it as described here. Also, don't forget to add the catalog to your album installation, so you can install the solutions from the catalog.
Install the CSBDeep-train solution by using the graphical user interface (GUI) of album or by running the following command in the terminal:
album install io.github.betaseg:CSBDeep-train:0.1.0
For prediction, install the CSBDeep-predict solution:
album install io.github.betaseg:CSBDeep-predict:0.1.0
How to use
CSBDeep-train
To run the training, set the parameters in the GUI or adapt this example for command line usage:
```bash
conda activate album
album run csbdeep_unet_train --root /data/csbdeep_unet_train/data_golgi --epochs 3 --steps_per_epoch 5
```
During training, a browser tab opens and shows the training progress. The program terminates after the training is finished and the tab is closed.
CSBDeep-predict
To perform inference/prediction, set the parameters in the GUI or adapt this example for command line usage:
```bash
conda activate album
album run csbdeep_unet_predict --input /data/csbdeep_unet_train/data_golgi/val/images --outdir /data/segmentations --model /models/2023_07_04-15_06_33_unet
```
For further options and default values, please refer to the corresponding info page of the solution:
album info csbdeep_unet_predict
Further documentation:
For further options, parameters and default values, please refer to the info page of the solution:
album info csbdeep_unet_predict
Hardware requirements
We recommend to use a GPU with at least 8GB of memory to run the solution as a minimum requirement.
Citation & License
This solution is licensed under the BSD 3-Clause License.
If you use this solution, please cite the following paper:
doi: 10.1038/s41592-018-0216-7,
title: Content-aware image restoration: pushing the limits of fluorescence microscopy